An international team
of scientists has sequenced and analyzed the genome of the African coelacanth, Latimeria chalumnae. Ancient lobe fins were the first
vertebrates to brave the land, and the coelacanth genome is expected to reveal
much about the origins of tetrapods, the evolutionary line that gave rise to
amphibians, reptiles, birds, and mammals. Coelacanths
have fleshy, lobed fins, that were believed to live during cretaceous period,
when dinosaurs still roamed earth. Ancient lobe fins were the first
vertebrates to brave the land. Coelacanth genome is expected to reveal much
about the origins of tetrapods, the evolutionary line that gave rise to
amphibians, reptiles, birds and mammals.
The findings were published in the magazine Nature, April
17, 2013.
http://www.scientificamerican.com/article.cfm?id=slow-evolving-lobe-finned-coelacanth-genome-unlocked&page=2
GENOME Research

Tuesday, January 5, 2021
African coelacanth: Whole Genome Sequence
Sunday, September 23, 2012
Custom gene editing rewrites zebrafish DNA
Researchers led by Stephen Ekker, a molecular biologist at the Mayo Clinic in Rochester, Minnesota, have for the first time made custom
changes to parts of the zebrafish (Danio rerio) genome, using
artificial enzymes to cut portions of DNA out of targeted positions in a
gene sequence, and replace them with synthetic DNA. The work is
published in Nature. Ekker and his colleagues are the first to use artificial TALENs for genome editing in live zebra fish larvae. TALENs have the potential to work on any DNA sequence and cause permanent modifications. They also allow faster analysis of
induced mutations — in some cases, the team found, it is possible to
observe effects in the injected larvae immediately.
Read more: Nature-doi:10.1038/nature.2012.11463
Saturday, December 31, 2011
Intronless genes in teleost fish genomes
A recent study made by German scientists revealed that
Takifugu rubripes, Tetraodon nigroviridis, Oryzias latipes, Gasterosteus
aculeatus and Danio rerio genomes are respectively comprised of 2.83%,
3.42%, 4.49%, 4.35% and 4.02% single exon genes (SEGs). These SEGs encode for a variety of
family proteins including claudins, olfactory receptors and histones
that are essential for various biological functions. Annotation features of three Dicentrarchus labrax chromosomes revealed 78 (5.30%)
intronless genes, comparisons with G. aculeatus showed that SEG
composition and their order varied significantly among corresponding
chromosomes, even for those with nearly complete synteny. More than half
of SEGs identified in most of the species have at least one ortholog
multiple exon gene in the same genome, which provides insight to their
possible origin by retrotransposition. In spite of the fact that they
belong to the same lineage, the fraction of predicted SEGs varied
significantly between the genomes analyzed, and only a low fraction of
proteins (4.1%) is conserved between all five species. Furthermore, the
inter-specific distribution of SEGs as well as the functional categories
shared by species did not reflect their phylogenetic relationships.
These results indicate that new SEGs are continuously and independently
generated after species divergence over evolutionary time as evidenced
by the phylogenetic results of single exon claudins genes. Results of this study provide strong support for the idea that retrotransposition
followed by tandem duplications is the most probable event that can
explain the expansion of SEGs in eukaryotic organisms.
Study was published in Marine Genomics. 2011 4(2):109-19.
Friday, September 23, 2011
First non-avian reptile whole genome sequence unveiled: Anolis carolinensis
Amniotes, the first truly terrestrial vertebrates, diverged from other animals some 320 million years ago to form the mammalian and reptilian lineages. Until now, however, the only representatives of the reptile branch to be sequenced were birds-the chicken, the turkey and the zebra finch.
Scientists from USA, UK and Sweden, recently reported the genome sequence of the North American green anole lizard, Anolis carolinensis. They found that A. carolinensis microchromosomes are highly syntenic with chicken microchromosomes, yet do not exhibit the high GC and low repeat content that are characteristic of avian microchromosomes. Comparative gene analysis shows that amniote egg proteins have evolved significantly more rapidly than other proteins. An anole phylogeny resolves basal branches to illuminate the history of their repeated adaptive radiations.
The genome sequence of A. carolinensis allows a deeper understanding of amniote evolution. Filling this important reptilian node with a sequenced genome has revealed derived states in each major amniote branch and has helped to illuminate the amniote ancestor.
The research is published in latest issue of Nature
Wednesday, September 21, 2011
Whole genome sequence: Anolis carolinensis lizard
Scientist found Anolis to have the most compositionally homogeneous genome of all amniotes sequenced thus far, a homogeneity exceeding that for the frog Xenopus. Isochores are large regions of relatively homogeneous nucleotide composition and are present in the genomes of all mammals and birds that have been sequenced to date. GC-rich isochores, with shorter introns and higher gene density are reported in all genomes sequenced till date, but disappeared from the Anolis genome. Using genic GC as a proxy for isochore structure so as to compare with other vertebrates, researchers found that GC content has substantially decreased in the lineage leading to Anolis since diverging from the common ancestor of Reptilia ∼275 MYA, perhaps reflecting weakened or reversed GC-biased gene conversion, a non-adaptive substitution process that is thought to be important in the maintenance and trajectory of isochore evolution.
Results demonstrate that GC composition in Anolis is not associated with important features of genome structure, including gene density and intron size, in contrast to patterns seen in mammal and bird genomes.
Findings are published in the latest issue of Genome Biology and Evolution
Labels:
amniotes,
anolis,
gene density,
introns,
isochores,
whole genome sequence
Alternative Splicing Switch
 |
| Adapted from Cell doi:10.1016/j.cell.2011.08.023 |
Alternative splicing (AS) is a key process underlying the expansion of proteomic diversity and the regulation of gene expression. Scientists from Canada and USA identified an evolutionarily conserved embryonic stem cell (ESC)-specific AS event that changes the DNA-binding preference of the forkhead family transcription factor FOXP1. An ESC-specific splicing switch in FOXP1 transcripts produces the FOXP1-ES isoform. FOXP1-ES has distinct DNA-binding properties compared to the canonical FOXP1 isoform. FOXP1-ES stimulates key pluripotency genes and represses many differentiation genes. FOXP1-ES is required for ESC pluripotency and efficient induced pluripotent stem cells (iPSC) reprogramming.
These results reveal a pivotal role for an AS event in the regulation of pluripotency through the control of critical ESC-specific transcriptional programs.
The findings are published in the recent issue of Cell
Mouse genomic variation
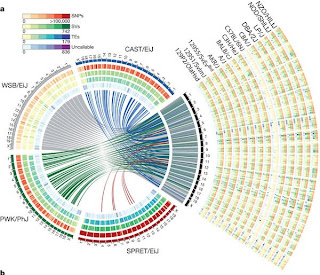 |
| Adapted from Nature 477, 289-294. |
Research outcomes are published in latest issue of Nature
Labels:
gene regulation,
Genomic variation,
phenotype
Tuesday, August 16, 2011
Whole genome Sequence of Atlantic Cod
Recently, scientists from Norway have investigated and present the genome sequence of Atlantic cod (Gadus morhua). The genome assembly was obtained exclusively by 454 sequencing of shotgun and paired-end libraries, and automated annotation identified 22,154 genes. Genome sequence provided evidence for complex thermal adaptations in its haemoglobin gene cluster and an unusual immune architecture compared to other sequenced vertebrates. Atlantic cod has lost the genes for MHC II, CD4 and invariant chain (Ii) that are conserved feature of the adaptive immune system of jawed vertebrates and, are essential for the function of this pathway. These observations affect fundamental assumptions about the evolution of the adaptive immune system and its components in vertebrates.
The study is published in the latest issue of Nature
Labels:
Atlantic cod,
CD4,
fish GENOMES,
Immune system,
Invariant chain (li),
MHC II,
whole genome sequence,
whole genome shotgun
Monday, April 18, 2011
Medaka Hd-rR: Whole Genome Sequencing Project
Sequencing of the medaka genome was started at the Academia Sequencing Center of the National Institute of Genetics (NIG) in mid 2002. The project was supported by group grant Genome Science (Grant-in-Aid for Scientific Research on Priority Areas supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan).
.
The sequencing was conducted by the whole-genome shotgun strategy using southern inbred strain, Hd-rR. The genome was assembled from 13.8 million reads, obtained from the whole genome shotgun plasmid, fosmid, and bacterial artificial chromosome (BAC) libraries. The total size of the assembled contigs was 700.4 megabases (Mb). 50% of nucleotides are covered in scaffolds (or contigs) of length 1.41Mb (9.8 kilobases) that are called N50 values. This contiguity is sufficient to characterize the genomic structures of genes.
Four versions of the medaka genome sequence data named 200406, 200506, version 0.9, and version 1.0 have been released to the public to provide users with timely information. The former two versions had shorter scaffolds that were not anchored on the medaka chromosomes because they were built in 2004 and 2005, before genetics markers were available. Versions 0.9 and 1.0 created in 2006, when comprehensive genetic markers were available, so that about 90% of their scaffolds and ultracontigs were located on the twenty-four medaka chromosomes. Versions 0.9 and 1.0 were built from the identical contigs and scaffolds, but the assembly of version 1.0 is longer than that of version 0.9 because more genetic markers could be used to generate version 1.0. Version 0.9 is left open to the public because most of the data analysis in the medaka genome paper published in Nature (2007) was based on version 0.9.
Labels:
BAC,
fish GENOMES,
fosmid,
Genome browser,
inbred starin,
medaka,
Nature,
plasmid,
whole genome sequence,
whole genome shotgun
Subscribe to:
Comments (Atom)





